Understanding the Coral Response

Motivation: Symbiosis is a complex and ecologically integrated interaction between organisms that provide emergent properties key to their survival. Such is the case for the relationship between reef-building corals and their microbiome, where nutritional and biogeochemical recycling provide the necessary benefits that fuel high reef productivity and calcification in oligotrophic waters. The pace of climate change is, however, taxing the capacity of coral reefs to adapt and maintain ecosystem function under increasing temperatures and ocean acidification. It is also unclear the extent to which traits of partners of the coral holobiont, the combined coral animal, its photosynthetic dinoflagellate endosymbionts (members of the Symbiodiniaceae), and associated prokaryotes, contribute to adaptive potential.

Specifically, a quantitative integration of holobiont function is not yet available. Furthermore, novel epigenetic mechanisms (i.e., DNA methylation and its link to gene expression) are coming to light in corals and add an additional layer of complexity to the holobiont stress response. Given these observations, the goals of this research project work are: 1) to characterize the host stress response to acute and chronic stressors in an environmentally resilient and a susceptible coral species, 2) to characterize the dynamics of the coral microbiome across stressors, treatments, and time, 3) to elucidate the role of the methylome in epigenetically regulating elements of stress response genes in corals, and 4) to quantify, using network analysis, the functional interactions between microbiome communities and host gene expression in experimental treatment conditions and link these data to the host methylome.
Major Partners: PIs: Hollie M. Putnam (University of Rhode Island), Ark Harel (Volcani Center, Israel), Xiaoyang Su (Rutgers University), Phillip Cleves (Stanford University), Cheong Xin Chan (University of Queensland, Australia), Hwan Su Yoon (Sungkyunkwan University, Korea), Tali Mass (University of Haifa, Israel), postdocs Jananan Pathmanathan, JunMo Lee, graduate students Amanda Williams, Julia Van Etten, Alexander Shumaker, staff researcher Eric Chiles, undergraduates George Soliman, Daniel Liu, Deeksha Misri.
Example Research Outcomes

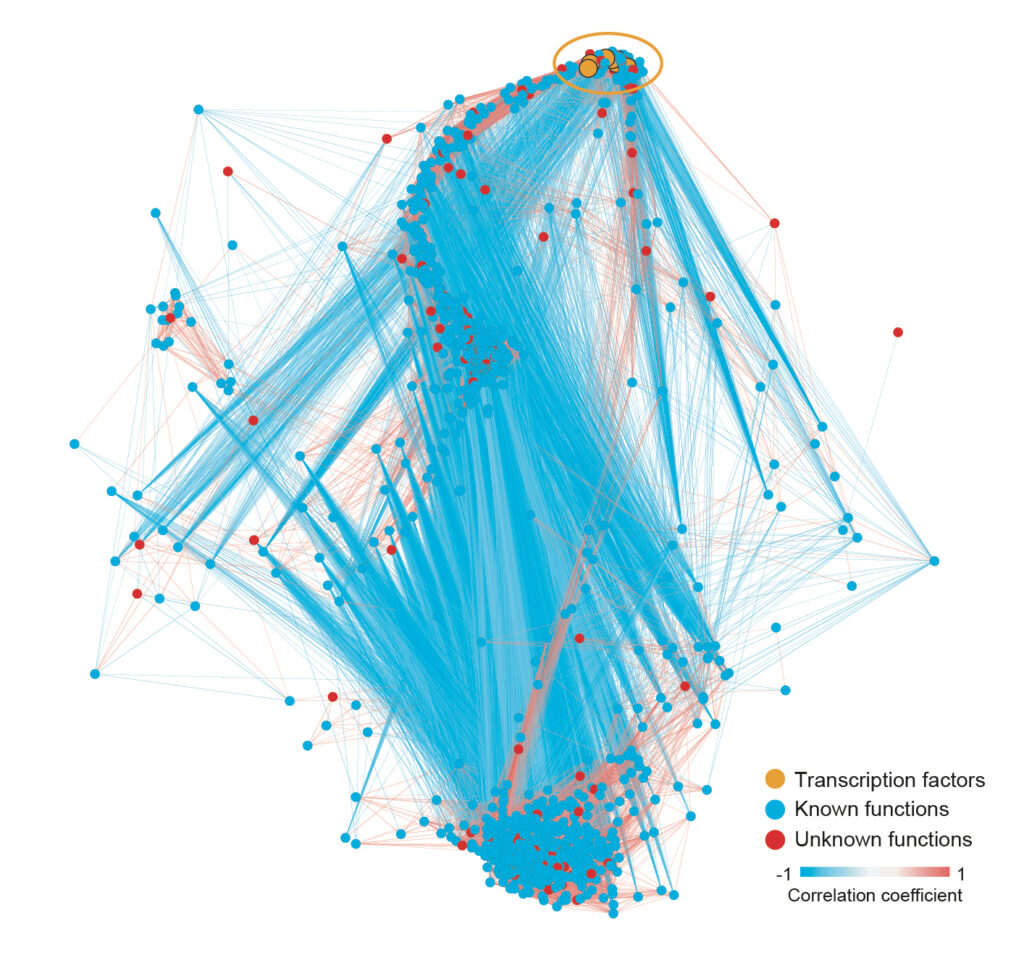
Representative Publications
- Van Etten J, Shumaker AK, Mass T, Putnam HM, Bhattacharya D. 2020. Transcriptome analysis provides a blueprint of coral egg and sperm functions. PeerJ [accepted].
- Shah S, Chen Y, Bhattacharya D, Chan CX. 2020. Sex in Symbiodiniaceae dinoflagellates: genomic evidence for independent loss of the canonical synaptonemal complex. Scientific Reports 10(1):9792.
- Cleves PA, Shumaker A, Lee J, Putnam HM, Bhattacharya D. 2020. Unknown to known: advancing knowledge of coral gene function. Trends in Genetics 36(2):93-104. (journal cover)
- Shoguchi E, Yoshioka Y, Shinzato C, Arimoto A, Bhattacharya D, Satoh N. 2020. Correlation between organelle genetic variation and RNA editing in finoflagellates associated with the coral Acropora digitifera. Genome Biology and Evolution 12(3):203-209.
- Palumbi SR, Anthony K, Baker AC, Baskett ML, Bhattacharya D, Bourne D, Knowlton N, Logan CA, Naish KA, Richmond RH, Smith TB, von Stackelberg K. 2019. National Academies of Sciences, Engineering, and Medicine. A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs. Washington, DC: The National Academies Press. https://doi.org/10.17226/25279.
- Shumaker A, Putnam HM, Qiu H, Price DC, Zelzion E, Harel A, Wagner NE, Gates R, Yoon HS, Bhattacharya D. 2019. Genome analysis of the rice coral Montipora capitata. Scientific Reports 9(1):2571.

