How Do Algae Become Extremophiles?

Motivation: Harsh environments with extremes of temperature, pH, solar radiation, salt concentration, and pressure produce exquisitely tailored solutions by resident biota. Bacteria and Archaea are the usual masters of life at the extreme, and until recently, algae that have evolved under similar conditions, have been largely overlooked. We focus here on the Cyanidiophyceae, a group of unicellular aquatic and terrestrial red algae (e.g., Galdieria, Cyanidium, and Cyanidioschyzon) that occupy a variety of hot springs and acid mining sites characterized by variable light levels, high temperature, low pH, with high salt and toxic heavy metal concentrations.
This lineage also includes a handful of secondarily mesophilic species (the so-called “Cave Cyanidium”) that are examples of reversion to the ancestral state. Recent work by our group has shown that the diverged Cyanidiophyceae (and many other algal/protist lineages) follow the “1% rule”; i.e., on average, approximately 1% of their gene inventory comprises prokaryote genes acquired via horizontal gene transfer (HGT). We have also demonstrated that a majority of HGT candidates encode proteins with functions related to “polyextremophily”, including metal and xenobiotic resistance/detoxification, cellular oxidant reduction, carbon metabolism, amino acid metabolism, osmotic resistance, and salt tolerance. These traits make Cyanidiophyceae ideal models for engineering resistance genes and pathways into commercially important algae and plants to protect them from environmental stresses such as drought and heavy metal contamination.
Other applications include alga-based production of compounds such as phycocyanin, floridosides, and glycogen, recovery of rare earth elements, and detoxification of heavy metals. Heterotrophic growth of many Galdieria species offers the opportunity for flexible photobioreactor design to minimize operating costs.
The goals of our ongoing work are to: 1) Generate long-read based reference assemblies and RNA-Seq data from many Cyanidiophyceae. 2) Do whole-genome bisulfite sequencing to detect cytosine methylation (5-methylcytosine; 5-meC). 3) Generate metabolomic data for chosen samples based on RNA-Seq evidence, under both positive and negative ionization modes. 4) Generate Illumina short-read metagenomic data for a time series at 3 distinct hot springs habitats at Lemonade Creek, Yellowstone National Park (YNP).
Major Partners: PIs: Timothy McDermott (Montana State University), Hwan Su Yoon (Sungkyunkwan University, Korea), Cheong Xin Chan (University of Queensland, Australia), Andreas PM Weber (Heinrich-Heine University), Na’ama Segal (Israel Oceanographic and Limnological Association), Xiaoyang Su (Rutgers University), graduate students Julia Van Etten.
Example Research Outcomes

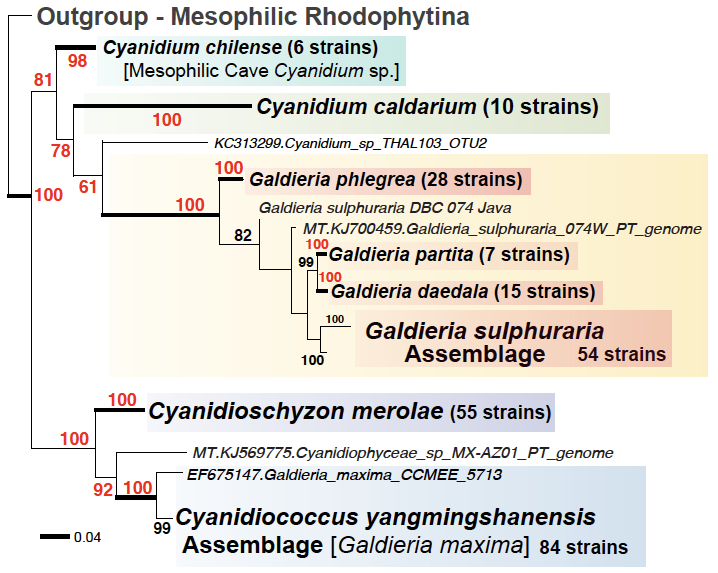

Representative Publications
- Wang D, Yu X, Xu K, Bi G, Cao M, Zelzion E, Fu C, Sun P, Liu Y, Kong F, Du G, Tang X, Yang R, Wang J, Tang L, Wang L, Zhao Y, Ge Y, Zhuang Y, Mo Z, Chen Y, Gao T, Guan X, Chen R, Qu W, Sun B, Bhattacharya D, Mao Y. 2020. Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nature Communications [accepted].
- Fan X, Qiu H, Han W, Wang Y, Xu D, Zhang X, Bhattacharya D, Ye N. 2020. Phytoplankton pangenome reveals extensive prokaryotic horizontal gene transfer of diverse functions. Science Advances 6(18):eaba0111.
- Zhang Z, Qu C, Zhang K, He Y, Zhao X, Yang L, Zheng Z, Ma X, Wang X, Wang W, Wang K, Li D, Zhang L, Zhang X, Su D, Chang X, Zhou M, Gao D, Jiang W, Leliaert F, Bhattacharya D, De Clerck O, Zhong B, Miao J. 2020. Adaptation to extreme Antarctic environments revealed by the genome of a sea ice green alga. Current Biology Jul 2: S0960-9822(20)30845-9. doi:10.1016/j.cub.2020.06.029.
- Rossoni AW, Price DC, Seger M, Lyska D, Lammers P, Bhattacharya D, Weber APM. 2019. The genomes of polyextremophilic Cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. eLife 8:e45017.
- Qiu H, Rossoni AW, Weber APM, Yoon HS, Bhattacharya D. 2018. Unexpected conservation of the RNA splicing apparatus in the highly streamlined genome of Galdieria sulphuraria. BMC Evolutionary Biology 18:41.
- Qiu H, Price DC, Yang EC, Yoon HS, Bhattacharya D. 2015. Evidence of ancient genome reduction in red algae (Rhodophyta). Journal of Phycology 51:624-636.
- Qiu H, Price DC, Weber AP, Reeb V, Yang EC, Lee JM, Kim SY, Yoon HS, Bhattacharya D. 2013. Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Current Biology 23:R865-866.

