Endosymbiotic Origin of Photosynthetic Organelles
Motivation: The Great Oxygenation Event (GOE) occurred ca. 2.4 billion years ago (Ga) and forced a major redox transition in the history of life. It was driven primarily by the spread of oxygenic photosynthesis in cyanobacteria and set in motion major changes in cellular metabolism and catalytic conversions in response to rising oxygen (O2) levels. Oxygenic photosynthesis entered the eukaryotic domain through cyanobacterial primary endosymbiosis over a billion years ago in the common ancestor of the eukaryotic “supergroup” Archaeplastida (red, green, and glaucophyte algae and land plants). The red and green algae gave rise, through subsequent serial endosymbioses, to a vast array of other unicellular and multicellular algae such as diatoms, dinoflagellates, and kelps that are critical to aquatic ecosystems and to global carbon cycles. The GOE and increasing prominence of photosynthetic eukaryotes created conditions that favored the evolution of complex, multicellular life. Specifically, the use of O2 as a terminal electron acceptor in respiration greatly increased the energy yield of glycolysis, and oxygenic photosynthesis fostered the evolution of multicellular photosynthetic organisms and colonization of non-aquatic environments by emerging advanced life forms.

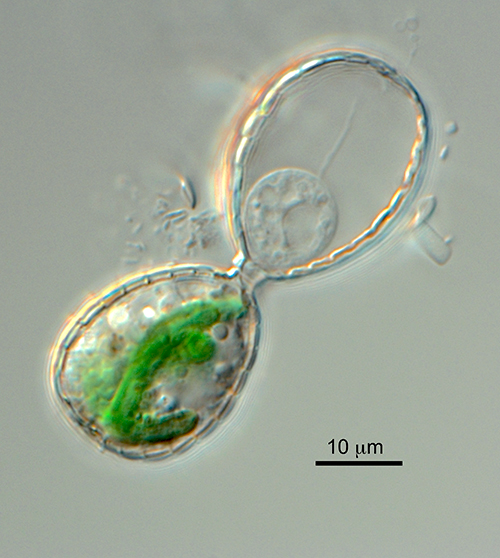
Plastid endosymbiosis is undoubtedly one of the most important innovations in the history of our planet. Therefore, a massive effort has been expended to elucidate plastid origin, function, biogenesis, diversity and its impacts on global primary productivity and geochemical cycles. What past work on members of the Archaeplastida and other algal models cannot provide however, due to the ancient derivation of their plastid, are insights into earlier stages of organellogenesis. During this critical phase, biotic interactions and genetic innovations evolved to stabilize, integrate and regulate the association between the host and the O2-evolving, potentially toxic (because of the production of reactive O2 species [ROS]) endosymbiont that was compromised by gene loss due to Muller’s ratchet. In this project, we fill a wide gap in knowledge of the evolution of early life by studying the origin of primary plastids in the only non-Archaeplastida model available. This model is a thecate amoeba (rhizarians), Paulinella, which acquired its plastid (referred to as the chromatophore) ca. 100 million years ago from an a-cyanobacterium. Our goals are to: 1) using omics and physiology, generate an understanding of the light response in Paulinella micropora. 2) Generate a genome-scale metabolic network (GEM) model for P. micropora in order to gain a systems-level understanding of chromatophore integration. 3) Using a heterologous expression system (Synechocystis sp. PCC 6803, among others), to study the functions of P. micropora stress-related genes.
Major Partners: PIs: Arthur R. Grossman (Carnegie Institution, Stanford), Hwan Su Yoon (Sungkyunkwan University, Korea), Cheong Xin Chan (University of Queensland, Australia), Eva Nowack (Heinrich-Heine University, Germany), postdocs Timothy Stephens, JunMo Lee, graduate students Arwa Gabr, Julia Van Etten.
Example Research Outcomes

Representative Publications
- Gabr A, Grossman AR, Bhattacharya D. 2020. Paulinella, a model for understanding plastid primary endosymbiosis. Journal of Phycology Apr 14. doi:10.1111/jpy.13003.
- Lhee D, Ha J-S, Kim S, Park MG, Bhattacharya D, Yoon HS. 2019. Evolutionary dynamics of the chromatophore genome in three photosynthetic Paulinella species. Scientific Reports 9(1):2560.
- Bhattacharya D, Price DC. 2020. The algal tree of life from a genomics perspective. In: Photosynthesis in Algae: Biochemical and Physiological Mechanisms. Eds: Larkum AWD, Grossmann AR, Raven JA. Springer International Publishing. DOI 10.1007/978-3-030-33397-3_2.
- Lee J, Kim D, Bhattacharya D, Yoon HS. 2019. Expansion of phycobilisome linker gene families in mesophilic red algae. Nature Communications 10:4823.
- Price DC, Goodenough UW, Roth R, Lee J-H, Kariyawasam T, Mutwil M, Ferrari C, Facchinelli F, Ball SG, Cenci U, Chan CX, Wagner NE, Yoon HS, Weber APM, Bhattacharya D. 2019. Analysis of an improved Cyanophora paradoxa genome assembly. DNA Research 26(4):287–299.
- Ferrari C, Proost S, Janowski M, Becker J, Nikoloski Z, Bhattacharya D, Price DC, Tohge T, Bar-Even A, Fernie A, Stitt M, Mutwil M. 2019. Kingdom-wide comparison reveals the evolution of diurnal gene expression in Archaeplastida. Nature Communications 10:737.
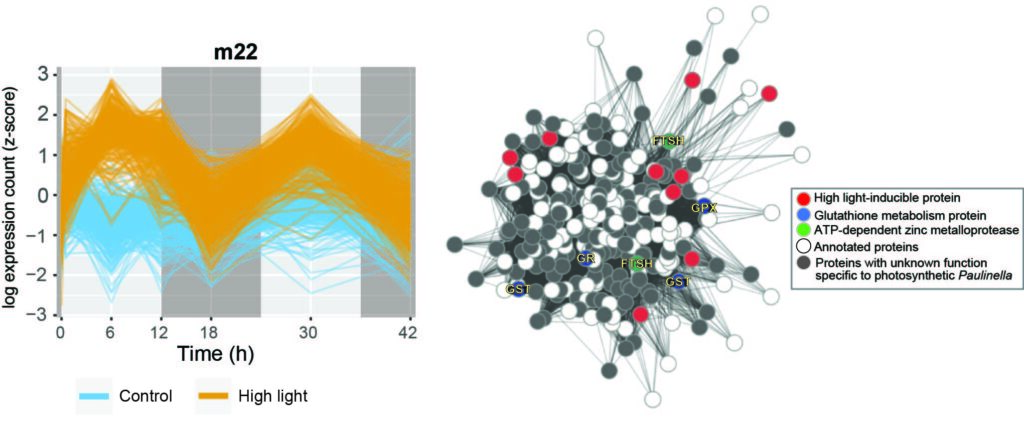
- Zhang R, Nowack ECM, Price DC, Bhattacharya D, Grossman AR. 2017. Impact of light intensity and quality on chromatophore and nuclear gene expression in Paulinella chromatophora, an amoeba with nascent photosynthetic organelles. Plant Journal 90(2):221-234.
- Nowack EC, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR. 2016. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proceedings of the National Academy of Sciences USA 113:12214-9.

